Points to note about GlycoCT
- Underdetermined capping unit
- Selection of residues connecting ambiguous antenna residues
- Change of substituent type
- Available substituents in GlycoCT
1. Underdetermined capping unit
- GlycoCT with the following glycan structure is using “u(-1+1)u” in GlycanBuilder. However, in the GlycoCT manual, as the type of identifier in the LIN section “u” is not described. Fig. 25 (p. 27) illustrates how to include an undetermined capping location. The notation used for the SubTreeLinkage is “o(4+1)d”. The linkage for the glycan structure G02402FF shown below should be represented as “o(-1+1)d”.
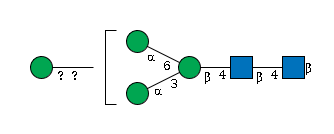
incorrect GlycoCT
Point: SubtreeLinkageID1:u(-1+1)u
RES
1b:b-dglc-HEX-1:5
2s:n-acetyl
3b:b-dglc-HEX-1:5
4s:n-acetyl
5b:b-dman-HEX-1:5
6b:a-dman-HEX-1:5
7b:a-dman-HEX-1:5
LIN
1:1d(2+1)2n
2:1o(4+1)3d
3:3d(2+1)4n
4:3o(4+1)5d
5:5o(3+1)6d
6:5o(6+1)7d
UND
UND1:100.0:100.0
ParentIDs:1|3|5|6|7
SubtreeLinkageID1:u(-1+1)u
RES
8b:x-dman-HEX-1:5
correct GlycoCT
Point: SubtreeLinkageID1:o(-1+1)d
RES
1b:b-dglc-HEX-1:5
2s:n-acetyl
3b:b-dglc-HEX-1:5
4s:n-acetyl
5b:b-dman-HEX-1:5
6b:a-dman-HEX-1:5
7b:a-dman-HEX-1:5
LIN
1:1d(2+1)2n
2:1o(4+1)3d
3:3d(2+1)4n
4:3o(4+1)5d
5:5o(3+1)6d
6:5o(6+1)7d
UND
UND1:100.0:100.0
ParentIDs:1|3|5|6|7
SubtreeLinkageID1:o(-1+1)d
RES
8b:x-dman-HEX-1:5
2. Selection of residues connecting ambiguous antenna residues
- Build glycan with a bracket using GlycoWorkbench (or GlycanBuilder)

- Export to GlycoCT format

- Find residues related to ambiguous antenna and their ID numbers in the GlycoCT

- Change the ID numbers specified at “ParentIDs” in the GlycoCT to select parent residues of ambiguous antenna
- Also need to change the connecting type in “SubtreeLinkageID”


3. Change of substituent type
- Build glycan with a substituent using GlycoWorkbench (or GlycanBuilder)
- The substituent must be able to convert to GlycoCT (e.g. phosphate)

- Export to GlycoCT format

- Find the substituent residue in the GlycoCT
- Change the substituent name to the other one which you want to add


4. Available substituents in GlycoCT

Available substituents in GlycoCT




